Rock Snot: Where It Comes From and How It's Made

This Behind the Scenes article was provided to LiveScience in partnership with the National Science Foundation.
Maybe you've heard the old joke: What's the worst thing you can do to a boat?
Put it in the water.
Once a boat enters the water, microorganisms begin accumulating on its surfaces, creating a significant amount of drag and a big mess. This may not matter for a small fishing boat, but for giant container ships, the drag created by microorganisms — in particular single-celled, silica-shelled organisms called diatoms — results in a significant amount of fuel loss every year.
Diatoms are one of the most common types of phytoplankton and a major group of algae. They constitute a large percentage of all living matter in oceans, rivers and lakes. One species, Didymosphenia geminata, is responsible for creating thick blooms in mountain streams and ponds. It is colloquially known as didymo or rock snot. The species is a menace to hospitals as well, where it can coat moist surfaces and promote bacteria by secreting mucilage that supports all types of growth.
For researchers in the lab of Edward Theriot at The University of Texas at Austin, diatoms (and their snot) are rich objects of biological research.
"There are hundreds of thousands of species out there in every body of water you can think of in the world," said Matt Ashworth, a researcher in the lab. "They're a very successful group, a relatively young group, and we're interested in understanding how they've evolved and colonized different ecosystems."
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
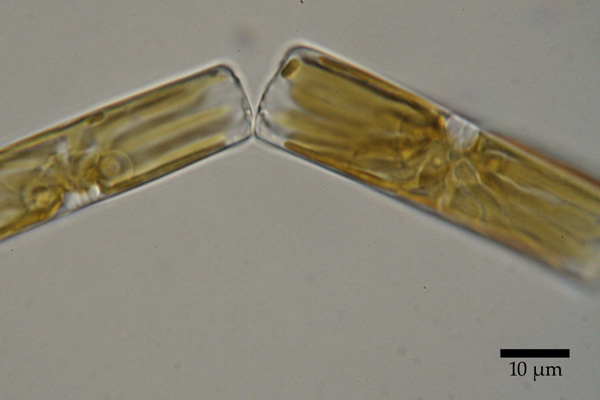
For decades, diatoms resisted study. Their genome is notoriously difficult to analyze. Morphological studies, based on the shape of a species' shell or other features, often contradicted the results of molecular testing. However, next-generation sequencing methods, in combination with computational techniques and powerful supercomputers, are helping researchers better understand the biology, evolution and dispersion of the diatom.
Using these new tools, researchers are trying to answer a number of basic questions about diatom evolution: What were the earliest diatoms like? How has the organism moved from a single site to every body of water in the world? And how have some species developed the ability to produce prodigious amounts of snot?

The Search for the Ur-Diatom
Researchers have sought to understand what the diatom's original ancestor might have looked like. Though Theriot and his team are still in the early stages of their analysis, some of their results are consistent enough that they can start to paint a picture of how the so-called "ur-diatom" may have appeared.
"There's this idea that the early diatom was a small flagellate, but what we're finding at the base of the diatom tree are things that are long and tubular, much like the tube inside of a paper towel roll," said Edward Theriot, professor of molecular evolution at The University of Texas at Austin and director of its Texas Natural Science Center. "In the diatom world, this is a radical view, but it's exactly what the tree is telling us."

To come to this conclusion, the researchers analyzed ribosomal and chloroplast genes of more than 200 diatoms and Bolidomonas (a closely related genus). They wanted to test the prevailing understanding of where certain diatoms fall on the evolutionary tree, and included species used as models in genomic studies and diatoms whose placement in the diatom phylogeny has been problematic or controversial.
After generating massive amounts of data using next-generation gene sequencers, they used the Ranger supercomputer at the Texas Advanced Computing Center to align, organize and analyze the DNA data, and to run phylogenetic programs that sketched out the evolution of diatoms as a whole.
"There are a number of programs that compare DNA sequences and estimate how those DNA sequences evolved from each other, and some of these have very complex algorithms," Ashworth said. "Before we had access to Ranger it would take weeks and months to run. Ranger does the same analyses in hours. So it's been a very powerful tool to give us quick ideas about how different strains are related to each other."
The time that Ranger saves the researchers doesn't just get them to an answer faster. It also allows them to test many alternative hypotheses.
"Sometimes, the best tree from molecular data looks radically different from what morphology tells us to expect," Theriot explained. "With Ranger, we can redirect our time away from just finding the best tree for a dataset, towards asking how different that best tree is from what scholars thought about diatom evolution, say, 100 years ago."
Using a statistical comparative approach, the researchers arrived at a different tree of diatom evolution than traditionally conceived, and a different point of origin. They reported some of their early findings at the XXII International Diatom Colloquium in August 2012 and they continue to investigate the sequencing results in light of previous research.


So how does the didymo make all that mucus?
For some of the lab's more focused studies, like the evolution of the rock snot's mucus-producing capability, the researchers sequenced the transcriptomes (all of the messenger RNA molecules expressed from an organism's genes) of half a dozen species to identify the key genes for the molecular production of the mucilage.
"People have been trying to characterize that mucilage chemically for 20 years and haven't come up with much information," Ashworth said. "Taking a transcriptome approach, I can generate a lot of data very quickly and tackle the issue not at the end results — which is the sugar that is secreted — but at the very beginning, at the point of the molecular machinery that assembled and allowed for the secretion of that sugar in the first place."
Four of the taxa they sequenced produce visible mucilage, and the other three taxa do not, but are closely related to the mucilage producers. They believe that closely related diatoms should share similar transcriptomes, except for the mucilage-related genes.
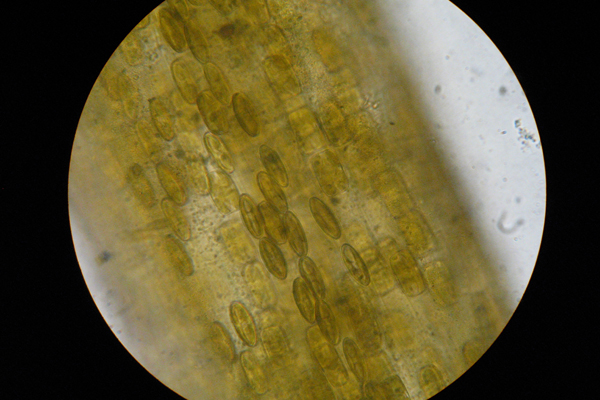
There are hundreds of genes involved in the assembly, packaging and secretion of these products, Ashworth said. If he can find 10 genes that are definitively involved in this process, then he is 10 genes closer to understanding how this function occurs.
"Generating DNA sequences in itself isn't particularly exciting, but the way the sequences fit together, or the existence of certain sequences at all, tells us a lot about the biology of these organisms."
Theriot uses TACC to host a web portal that supports the research in the lab, called Protist Central. He and his team use the portal to manage images and information about all the diatoms that they're working on. They also use it to manage information from their collaboration with researchers in Guam on the diatom flora of the coral reefs of the Pacific. Want to explore the beautiful and microscopic world of diatoms? Check out the image gallery, Diatoms of the Texas Gulf Coast.
Editor's Note: The researchers depicted in Behind the Scenes articles have been supported by the National Science Foundation, the federal agency charged with funding basic research and education across all fields of science and engineering. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author and do not necessarily reflect the views of the National Science Foundation. See the Behind the Scenes Archive.