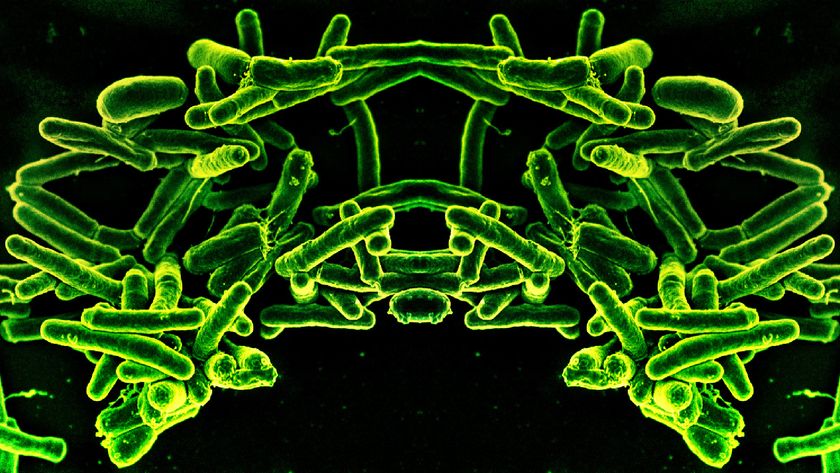
Bacteria Crafty at Sharing Antibiotic Resistance
Bacteria have more strategies for combating man-made antibiotics than previously thought, a new study suggests.
The results show that just a few antibiotic-resistant bacteria within a colony can confer protection to the entire bacterial community. But scientists might be able to exploit this protection strategy to develop treatments for antibiotic-resistant bacteria.
Previously, researchers thought each bacterium within a colony needed to individually develop antibiotic resistance. It did this either when it experienced a change in its genes, called a mutation, that conferred resistance, or when it picked up genes from other bacteria it came into contact with that provided resistance.
But the new study shows bacteria lucky enough to develop drug resistance are able to share their good fortune over longer distances with their neighbors, without changing their genes. Antibiotic-resistant bacteria can send out a signaling molecule that turns on protective mechanisms within the non-resistant bacteria, enabling them to survive in the presence of the drug.
"It indicates that these unicellular organisms can behave more or less as a multicellular organism," said study author James J. Collins, a researcher at the Howard Hughes Medical Institute in Boston.
The study is the first to show that signaling molecules can play a role in antibiotic resistance, Collins said. A better understanding of this type of bacterial communication may be critical for figuring out ways to prevent resistance, the researchers say.
The work will be published in the Sept. 2 issue of the journal Nature.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
Long-distance resistance
Recent years have seen a rise in the number of cases of antibiotic resistant bacteria, or "superbugs," including methicillin-resistant Staphylococcus aureus, or MRSA.
Collins and his colleagues were interested in learning how bacteria acquire the genetic mutations that provide resistance. They grew Escherichia coli in a bioreactor, an instrument that lets the researchers tightly control the environment the microbes are exposed to.
They then subjected the bacteria to increasing doses of the antibiotic norfloxacin. Every so often, they removed small samples of the bacterial population to test what is known as the "minimum inhibitory concentration" (MIC) — the minimum dose of an antibiotic that will prevent bacterial growth. Bacteria with higher MICs have better antibiotic resistance.
The researchers were surprised to see that most of their small samples had lower MICs than the population as a whole. But every so often, they stumbled across a sample that had a much higher MIC than the overall group.
Further research suggested that less than 1 percent of the bacterial population was actually resistant to the norfloxacin. These resistant bacteria aren't stressed out by the drug, and are able to help out the rest of the population by releasing a protein called indole into their environment.
"Indole enhances resistance of more susceptible cells, enabling them to survive and thrive in the face of the antibiotic even though they don’t necessarily have a mutation that affords them resistance to the antibiotic," Collins told MyHealthNewsDaily.
Indole switches on pumps inside the otherwise vulnerable bacteria, allowing them to expel the antibiotic. It also triggers pathways that protect the bacteria against free radicals — molecules with extra electrons that can cause damage to the bacteria. One of the main ways antibiotics are thought to kill bacteria is by bombarding them with free radicals.
By sending out indole, the antibiotic resistant bacteria seem to be acting altruistically. They gain no benefit; in fact, it is costly for these bacteria to produce indole. But by making this protein, they are helping out others that share their genes.
Future research
Targeting the pathway bacteria use to make indole might be a useful way to block the development of antibiotic resistance, Collins said. The researchers also found that E. coli produce indole in the presence of other antibiotics besides norfloxacin. They think this might be a widely used strategy that bacteria employ to share resistance, Collins said, but future work will be needed to see whether this is true.
Future studies will also investigate whether other molecules besides indole play a role in this sharing of antibiotic resistance.
The study was funded by the National Institutes of Health, the National Science Foundation and the Howard Hughes Medical Institute.
- 7 Solid Health Tips That No Longer Apply
- Could Humans Live Without Bacteria?
- Rise of Deadly Superbugs should 'Raise Red Flags' Everywhere
This article was provided by MyHealthNewsDaily, a sister site to LiveScience.

Rachael is a Live Science contributor, and was a former channel editor and senior writer for Live Science between 2010 and 2022. She has a master's degree in journalism from New York University's Science, Health and Environmental Reporting Program. She also holds a B.S. in molecular biology and an M.S. in biology from the University of California, San Diego. Her work has appeared in Scienceline, The Washington Post and Scientific American.