DeepMind AI has discovered the structure of nearly every protein known to science
The DeepMind program AlphaFold predicted the structures.
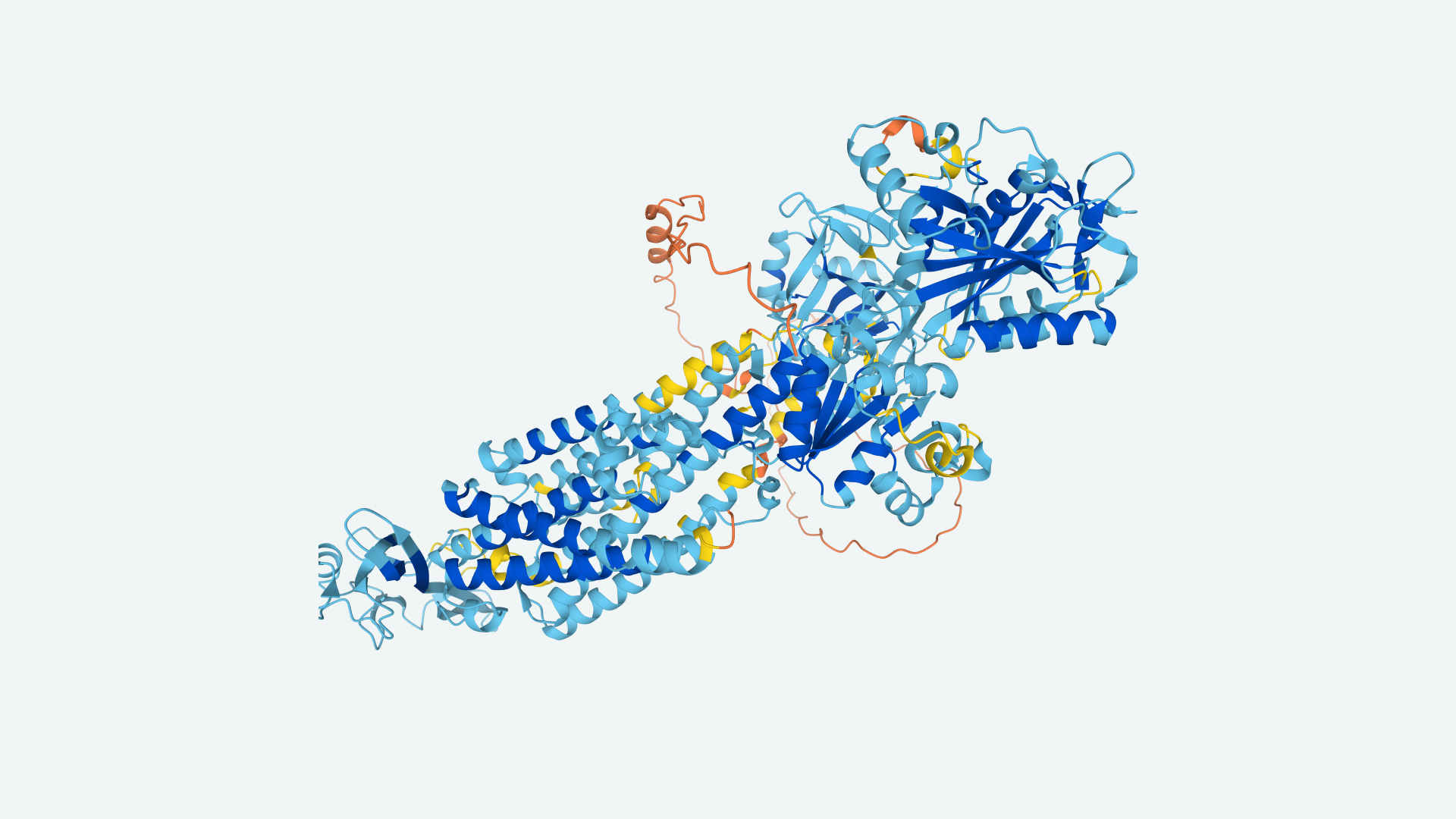
The artificial intelligence group DeepMind has unraveled the structures of nearly every protein known to science.
Researchers achieved the feat using the program AlphaFold, which DeepMind first developed in 2018 and released publically in July 2021. The open-source program can predict a protein's 3D structure from its sequence of amino acids, the building blocks that make up proteins. A protein's structure dictates its functions, so the database of 200 million protein structures identified by AlphaFold has the potential to help identify new protein workhorses that humans can make use of.
For example, the database may include proteins that can assist in recycling plastics, according to The Guardian.
"It took us quite a long time to go through this massive database of structures, but [it] opened this whole array of new three-dimensional shapes we’d never seen before that could actually break down plastics," John McGeehan, a professor of structural biology at the University of Portsmouth in the U.K., told The Guardian. "There’s a complete paradigm shift. We can really accelerate where we go from here — and that helps us direct these precious resources to the stuff that matters."
Deep dive into proteins

Proteins are like tiny, inscrutable puzzles. They are produced by organisms ranging from bacteria to plants to animals, and when they are made they fold up in milliseconds, but their structures are so complex that trying to guess what shape they'll take is nearly impossible. Cyrus Levinthal, an American molecular biologist, pointed out the paradox that proteins fold so quickly and precisely despite having huge numbers of possible configurations in a paper in 1969, estimating that a given protein might have 10^300 possible final shapes..
Thus, Levinthal wrote, if one tried to get to the correct protein shape by trying out each configuration one by one, it would take longer than the universe has existed so far to get to the right answer.
Scientists do have ways to visualize proteins and analyze their structures, but this is slow and difficult work. The most common way to image proteins is through X-ray crystallography, according to the journal Nature, which involves beaming X-rays at solid crystals of proteins and measuring how those rays are diffracted to determine how the protein is arranged. This experimental work had established the shape of about 190,000 proteins, according to DeepMind.
Get the world’s most fascinating discoveries delivered straight to your inbox.
Last year, DeepMind released protein shape predictions for every protein in the human body and in 20 research species, Live Science previously reported. Now, they've expanded those predictions to proteins in basically everything.
"This update includes predicted structures for plants, bacteria, animals and other organisms, opening up many new opportunities for researchers to use AlphaFold to advance their work on important issues, including sustainability, food insecurity and neglected diseases," DeepMind representatives said in a statement.
Making proteins work
AlphaFold works by accruing knowledge about amino acid sequences and interactions as it attempts to interpret protein structures. The algorithm can now predict protein shapes in minutes with accuracy down to the level of atoms.
Researchers are already using the fruits of AlphaFold's labor. According to The Guardian, the program enabled researchers to finally characterize a key malaria parasite protein that hadn't been amenable to X-ray crystallography. This, the researchers told The Guardian, could improve vaccine development against the disease.
At the Norwegian University of Life Sciences, honeybee researcher Vilde Leipart used AlphaFold to reveal the structure of vitellogenin —a reproductive and immune protein that is made by all egg-laying animals. The discovery could lead to new ways to protect important egg-laying animals like honeybees and fish from disease, Leipart wrote in a blog post for DeepMind.
The program is also informing the search for new pharmaceuticals, Rosana Kapeller, CEO of ROME Therapeutics, said in the DeepMind statement.
"AlphaFold speed and accuracy is accelerating the drug discovery process," Kapeller said,
"and we’re only at the beginning of realizing its impact on getting novel medicines to patients faster."
Originally published on Live Science.

Stephanie Pappas is a contributing writer for Live Science, covering topics ranging from geoscience to archaeology to the human brain and behavior. She was previously a senior writer for Live Science but is now a freelancer based in Denver, Colorado, and regularly contributes to Scientific American and The Monitor, the monthly magazine of the American Psychological Association. Stephanie received a bachelor's degree in psychology from the University of South Carolina and a graduate certificate in science communication from the University of California, Santa Cruz.
