188 new types of CRISPR revealed by algorithm
Researchers used an algorithm to scour databases of bacterial genomes for never-before-seen CRISPR systems.
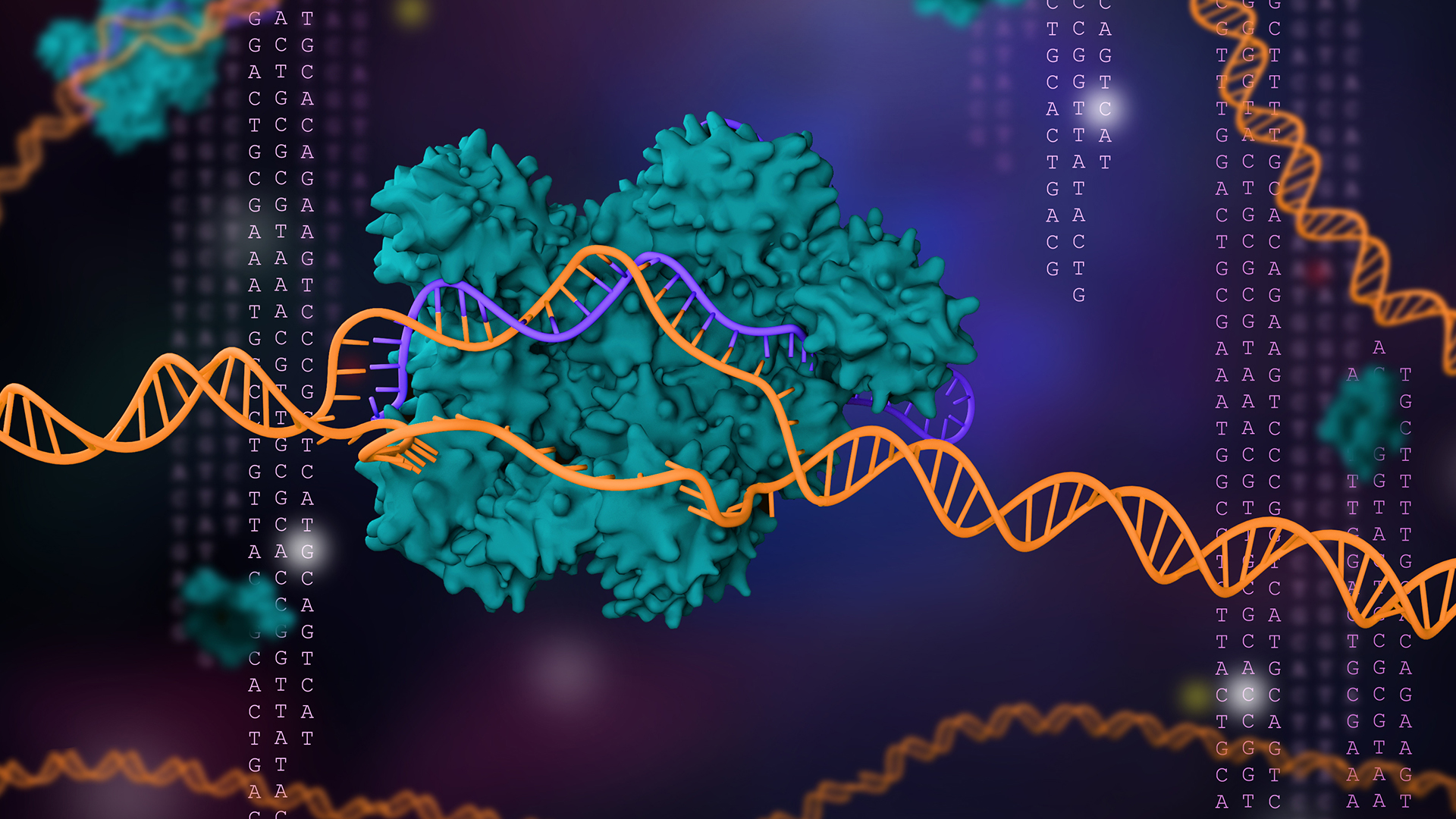
Scientists have unearthed 188 previously unknown types of CRISPR systems buried in the genomes of simple microorganisms.
Best known as a powerful gene-editing tool, CRISPR actually comes from an inbuilt defense system found in bacteria and simple microbes called archaea. CRISPR systems include pairs of "molecular scissors" called Cas enzymes, which allow microbes to cut up the DNA of viruses that attack them. CRISPR technology takes advantage of these scissors to cut genes out of DNA and paste new genes in.
The new study, published Nov. 23 in the journal Science, expands the known diversity of CRISPR systems in microorganisms and could open new avenues for precise gene editing with fewer "off-target" effects, the researchers said.
The team discovered the new CRISPR systems by scanning millions of genomes of microorganisms using an algorithm called fast locality-sensitive hashing-based clustering, or FLSHclust (pronounced "flash clust"). The algorithm works by very efficiently grouping similar objects together and is designed to hunt down genes related to CRISPR. The researchers used FLSHclust on three massive public datasets that contain billions of DNA and protein sequences from bacteria.
Related: The world's 1st CRISPR therapy has just been approved. Here's everything you need to know
"This new algorithm allows us to parse through data in a time frame that's short enough that we can actually recover results and make biological hypotheses," co-first study author Soumya Kannan, formerly a doctoral student and now a postdoctoral scholar in the Broad Institute/MIT Department of Biology, told MIT News.
FLSHclust accomplishes in weeks what previous algorithms would achieve in months, MIT News reported.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
After finding the new types of CRISPR, the researchers experimented with four of the systems to start understanding how they work.
The CRISPR systems that scientists previously knew about came in six flavors — types I through VI — which differ in size, the enzymes they use and how they latch onto genetic material, Nature reported. (The first CRISPR system ever identified is a type II system and uses a single enzyme called Cas9 to cut through DNA, while other types use different or multiple Cas enzymes.)
Out of the four CRISPR clusters the team experimented with, two were variants of type I CRISPR systems and one was type IV. Both type I systems made small, precise cuts in DNA in human cells, the team demonstrated. Scientists think that type I systems could potentially be less prone to making accidental, off-target cuts than CRISPR-Cas9, so they could be useful for gene editing, according to MIT News.
The final cluster was a whole new type of CRISPR, which the team dubbed type VII. Like some other CRISPR types, it targets RNA, a molecular cousin of DNA that's key for building proteins. So in theory, type VII systems could be useful for RNA editing.
That said, it's too soon to say whether type VII CRISPR systems or any of the other genes identified by FLSHclust will be helpful for genetic engineering, co-first study author Han Altae-Tran, formerly a graduate student at Broad Institute/MIT Department of Biology and now a postdoctoral scholar at the University of Washington, told Nature.
The next step will be to sift through more of the newfound systems to see how their component parts work and whether they could feasibly be used in gene editing, Nature reported.
Read more in MIT News and Nature.
Ever wonder why some people build muscle more easily than others or why freckles come out in the sun? Send us your questions about how the human body works to community@livescience.com with the subject line "Health Desk Q," and you may see your question answered on the website!

Nicoletta Lanese is the health channel editor at Live Science and was previously a news editor and staff writer at the site. She holds a graduate certificate in science communication from UC Santa Cruz and degrees in neuroscience and dance from the University of Florida. Her work has appeared in The Scientist, Science News, the Mercury News, Mongabay and Stanford Medicine Magazine, among other outlets. Based in NYC, she also remains heavily involved in dance and performs in local choreographers' work.