Ancient DNA and modern genomes can reveal stories of past peoples, from the Iron Age to Chernobyl, geneticist says
Ingrida Domarkienė, a geneticist at Vilnius University in Lithuania, discusses the exciting developments made possible by studying ancient and modern DNA.

Ingrida Domarkienė studies ancient DNA, weaving together fragments of genetic material from modern humans and our long-extinct human relatives to retell their stories.
From a background in molecular biology and medical genetics, Domarkienė now spearheads Lithuania's first ancient DNA lab, headquartered at the Medical Science Centre at Vilnius University. Along with international collaborators, the lab is studying the remains of people in medieval mass graves in Poland to learn about social practices that were prevalent in the region at the time, as well as the migrations of Iron Age individuals in Lithuania.
They're also revealing insights into the aftermath of the 1986 disaster at the Chernobyl nuclear power plant. Looking at the DNA of Lithuanian workers involved in the cleanup after the disaster, the researchers identified genes that help protect against the effects of radiation.
Live Science spoke with Domarkienė, who is also an associate professor at Vilnius, asking about her research, the unique challenges associated with studying ancient DNA, and how delving into our genetic history can lead to medical advances today.
Related: Modern Japanese people arose from 3 ancestral groups, 1 of them unknown, DNA study suggests
Emily Cooke: What is it about ancient DNA that you find interesting?
Ingrida Domarkienė: It's fascinating, how you can reassemble stories from DNA pieces, you know: You just sequence DNA; it's kind of the technological thing.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
And for me, coming from molecular biology, it's so fascinating that you read the biochemical fragment, organic molecule, and then you compare it with other samples, and you get a picture of how people moved, where they came from, where they went, how they lived. You can get "admixture signals" — that means that people [from different populations] mixed, and you can get an idea of who met whom and how they went on and on, and you can retell [their] stories.
EC: What are the unique challenges associated with studying ancient DNA?
ID: The most critical challenge is that you have to embrace uncertainty and failure here. Why is that? Because you are never quite sure if you will get the quality and quantity of DNA to work on further.
That's because, when an organism dies, DNA starts to decay, and there's nothing there to repair the DNA as it is in living cells. So it starts to fragment, and changes in composition. What's more, it blends with all other environmental DNA, which, when extracted, appears as contamination. So, in this case, I like an analogy of a confetti — or what is left of it after a huge celebration.
EC: Can you talk a bit about your research on Chernobyl survivors?
ID: Chernobyl survivors — cleanup workers or liquidators, they're also called.
It was our project with colleagues at the Department of Human and Medical Genetics, and in the group, we had the idea to analyze the genomes of Chernobyl liquidators, and we invited them to participate in the study. And when they started coming, we heard their stories, and we understood that — you know, those people went through a lot, but still, there were so many of them who were aging quite healthy, without cancers. You could expect the worst outcomes after what they've been through, but they were quite OK.

And then we got this hypothesis: that maybe there is something in the genomes of those survivors that protects them from all that bad that happened, let's say — also, psychological stress, which was immense back then, when they were taken from where they were at the moment and brought to Chernobyl without saying a word. They were telling stories of how they were woken up, and they were just there on a train going to God knows where.
And then that was a hard time, of course, there, and they had not only to work hard through liquidation work, but also trying to keep sane in that kind of place.
So, we started analyzing their genomes, and we found some potential signals of protective variation. And then we also have this new [as of yet unpublished] paper written by our student, which is on mitochondrial DNA. So, those Chernobyl liquidators might have protective variants also in the mitochondrial genome and nuclear genome [DNA in the nucleus] that supports mitochondrial function. So perhaps that's the idea.
Related: Chernobyl's liquidators didn't pass on radiation damage to their children
EC: How can studying our genetic history help us address medical challenges today?
ID: Studying the past through ancient DNA is fundamental research and takes time to realize what the findings are and how they can be implemented in practice. Perhaps the most important ingredient would be a strong interdisciplinary team that you can trust, and you can't do anything alone, really.
I find Svante Pääbo's work as a benchmarking example: How can you talk about ancient DNA without mentioning Svante's name?
He and his team developed the whole field of paleogenomics and generated the reference genome of Neanderthals. They started to give us explanations of what the differences between human and Neanderthal sequences are and what they mean in a functional way. For example, one of Svante Pääbo's group scientists, Dr. Hugo Zeberg, with colleagues, found that the Neanderthal variant in the progesterone receptor is associated with preterm birth, but also protective against miscarriage, and results in more live births. That knowledge can be translated into real help for women to save their pregnancies.
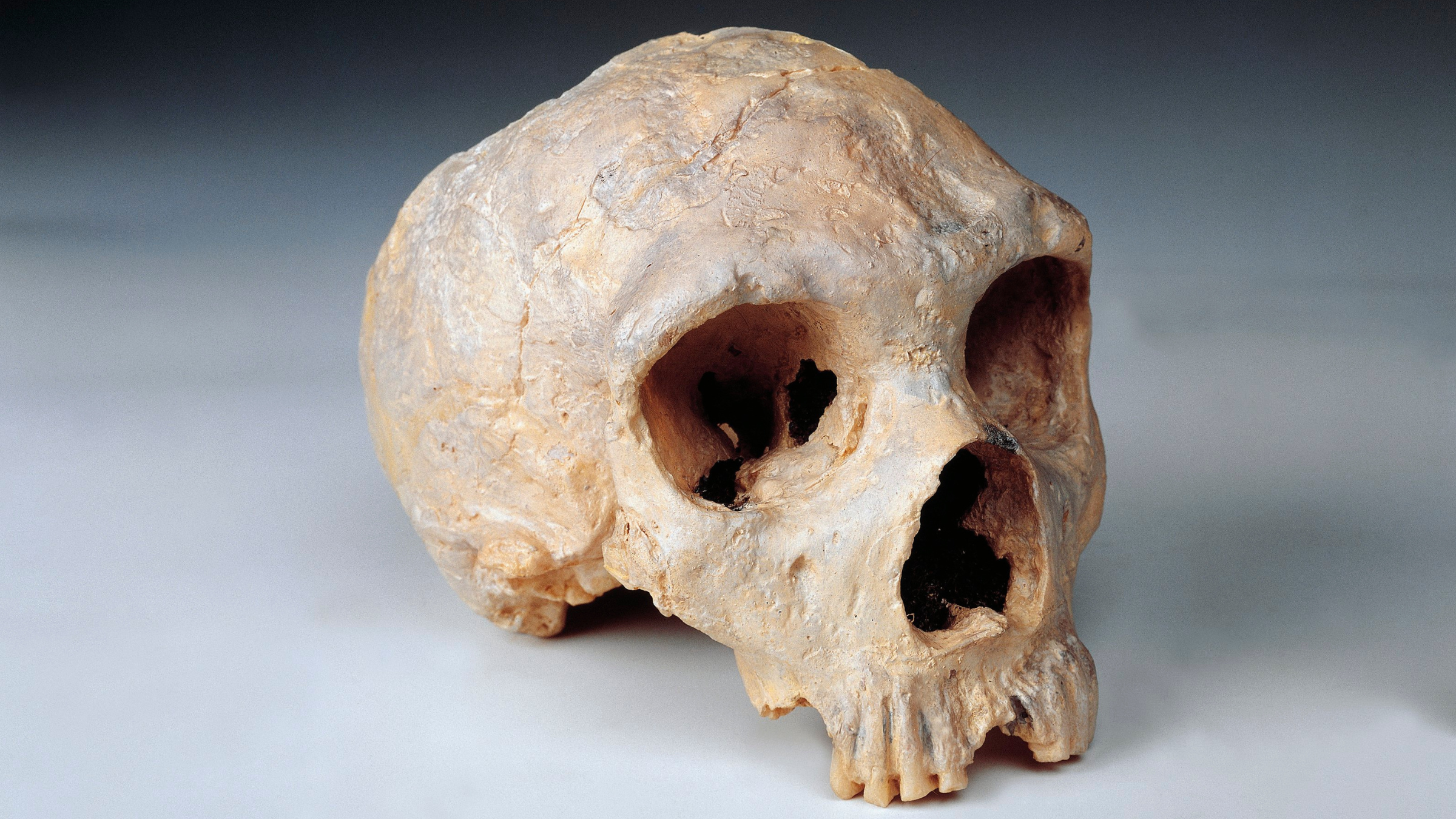
Or, another love story: metagenomics [the study of genetic material from all organisms in an environment], which is [an] even more challenging field. But it can help with infectious diseases, as we just witnessed one of the pandemics — and with the changing climate, there might be even more. So while reconstructing genomes of pathogens and building phylogenetic trees [diagrams of evolutionary relationships between species], we can understand the ways pathogens evolve and spread.
With those analyses, we can even start new narratives. For example, a long time ago, it was thought that the Spaniards introduced tuberculosis to the New World. But professor Johannes Krause's team [at the Max Planck Institute for Evolutionary Anthropology] showed that the bacteria was there before Columbus was, and apparently it was brought and transferred to humans by seals, which were a nutritious food for the people living there in Peru.
So you can see that, with this field, we can give science and medicine even more.
EC: What do you think the future of ancient DNA research will look like?
ID: From my point of view, I think that with the fast-evolving technologies, we will be able to go deeper in the sequences, wider in the datasets, and more divergent markers that we analyze.
Because now, we usually analyze single nucleotide polymorphisms, or SNPs [pronounced "snips," which are variations in single building blocks of DNA], as we call them. But my dream would be to reconstruct copy number variation, which are the huge chunks of [repeated] DNA, and it's not possible to do that now, but there are initiatives to do that.
We can also go for analysis of epigenomic markers [changes to DNA across the genome that alter the activity of genes without affecting the underlying sequence], which are very good markers for analysis of how the genome is regulated — to understand how it was back then. Those epigenomic markers also would be of great value.
And besides analysis of sociocultural structures of ancient sites, I would say research will definitely be directed towards understanding the functional meaning of the DNA variation that we analyze.
And in the grand finale, we would be integrating the whole data of the holobiome. That means all genomic information from the environment — not only humans, but also bacteria, viruses, plants, animals, everything who lives there. And integrating this data not only from the different disciplines, but also with different methods we have. Because the data comes using different methods, and that would be nice to integrate everything. And maybe then we will have the complete picture.
Editor's note: This interview has been condensed and edited for clarity.

Emily is a health news writer based in London, United Kingdom. She holds a bachelor's degree in biology from Durham University and a master's degree in clinical and therapeutic neuroscience from Oxford University. She has worked in science communication, medical writing and as a local news reporter while undertaking NCTJ journalism training with News Associates. In 2018, she was named one of MHP Communications' 30 journalists to watch under 30. (emily.cooke@futurenet.com)
You must confirm your public display name before commenting
Please logout and then login again, you will then be prompted to enter your display name.