Oldest sequenced DNA belonged to 1 million-year-old mystery mammoth
DNA from three different mammoths revealed ice age secrets.

The oldest DNA ever decoded belonged to a mammoth from a mysterious, previously unknown lineage that lived about 1.2 million years ago, a new study finds.
Previously, the oldest known sequenced genome came from a horse that lived up to 780,000 years ago, in what is now Canada's Yukon Territory. Now, the mammoth discovery, "is, with a wide margin, the oldest DNA ever recovered," study senior researcher Love Dalén, a professor of evolutionary genetics at the Center for Palaeogenetics in Stockholm, said at a news conference Tuesday (Feb. 16).
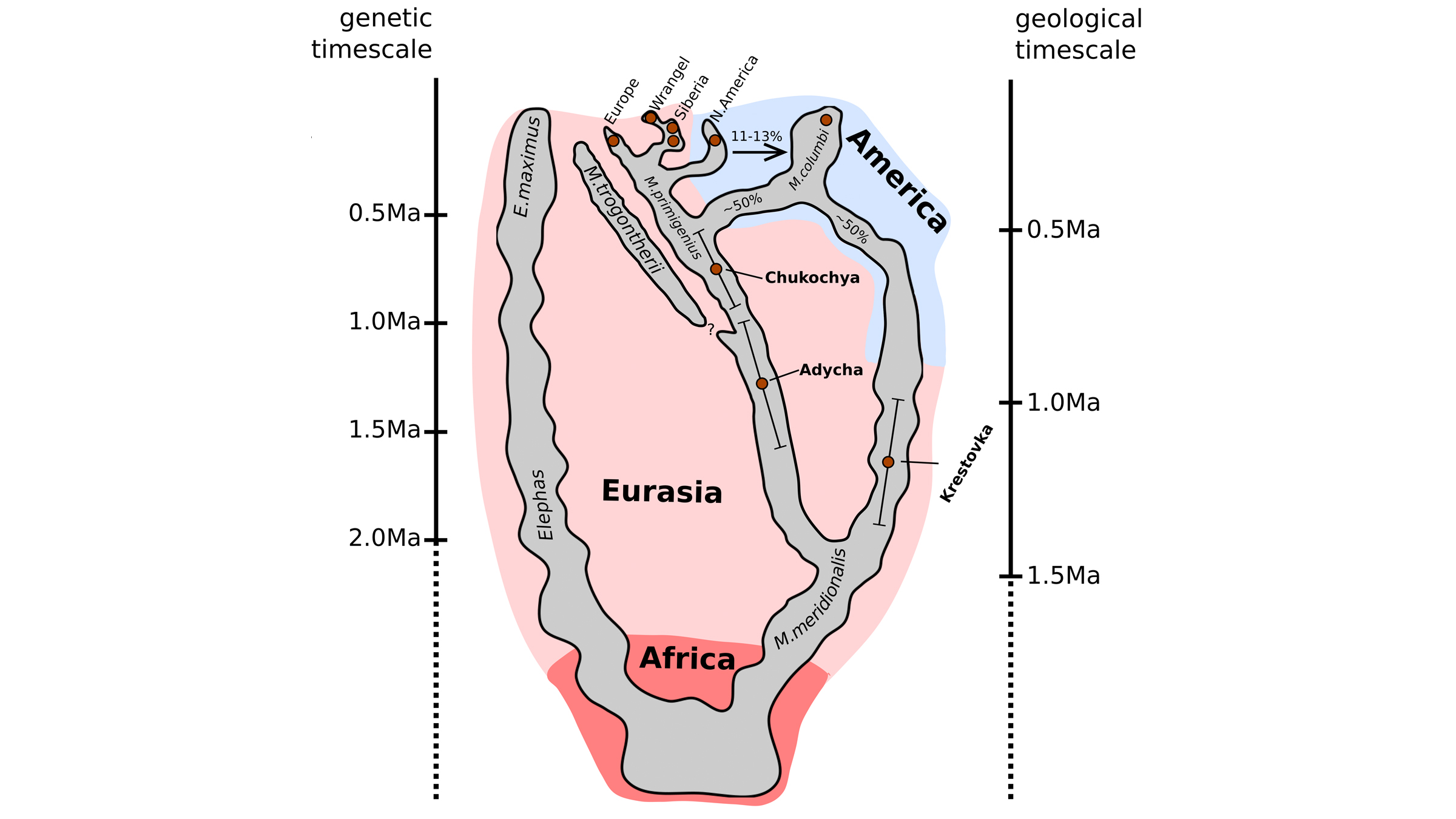
The remains of the mysterious mammoth were discovered near Siberia's Krestovka River (now the mammoth's namesake). After studying its ancient DNA, along with the newly sequenced genomes of two other mammoths — a roughly 700,000-year-old woolly mammoth (Mammuthus primigenius) and an approximately 1 million-year old woolly mammoth predecessor — the scientists made a surprising discovery: Woolly mammoths mated with a mammoth from Krestovka's mysterious line about 420,000 years ago, leading to a hybrid mammoth we know today as the Columbian mammoth (Mammuthus columbi).
Related: Mammoth resurrection: 11 hurdles to bringing back an ice age beast
The Krestovka mammoth lineage, the researchers found, would have migrated from Siberia to North America, crossing over the Bering Strait land bridge, about 1.5 million years ago; fossil evidence suggests that the woolly mammoth crossed over about 100,000 years ago, but the researchers say it could have come earlier, perhaps 500,000 to 400,000 years ago.
It's unknown where the hybridization occurred, but when they met up, "these two lineages hybridized and formed what we now call the Columbian mammoth," lead researcher Tom van der Valk, a postdoctoral researcher at the Swedish Museum of Natural History and the Center for Palaeogenetics, said at the news conference.
The genetic discovery of the Columbian mammoth's origins is remarkable, said Daniel Fisher, a professor and curator in the Museum of Paleontology of the University of Michigan, who was not involved with the study. "Hybrid origins of new species have certainly been proposed before, but few cases have placed the phenomenon in such a clear historical, geographic and evolutionary context," Fisher told Live Science in an email.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.




Woolly project
When the researchers began the project, they hoped to learn about woolly mammoth evolution — "Our main aim was to recover genomes from before and after the origin of the woolly mammoth," Dalén said. They chose to study three mammoths whose remains had been found in northeastern Siberia by the late Russian paleontologist Andrei Sher in the 1970s. These included one of the oldest woolly mammoths on record, the 700,000-year-old pachyderm nicknamed Chukochya, and the woolly mammoth predecessor, called Adycha, which looked like a European steppe mammoth (Mammuthus trogontherii), a direct ancestor of the woolly mammoth.
The third mammoth, Krestovka, also looked a bit like a steppe mammoth, even though it had diverged from that lineage more than 2 million years ago. So, despite evolving on entirely different lineages for more than 1 million years, the mammoths from the woolly mammoth and Krestovka lines were still able to mate, the researchers found.
"It looks like hybridization of lineages that were long separated, on the order of 1 million years, [were] coming together and successfully producing an apparently new species, which is really mind-blowing," said Ross MacPhee, a senior curator of vertebrate zoology at the American Museum of Natural History in New York City, who was not involved with the study.
By studying woolly mammoth genetics, the researchers made another discovery: "Many of the adaptations that we know woolly mammoths had, such as thermoregulation, changes in their circadian rhythm, fat deposits and hair growth were already present in the steppe mammoth," van der Valk said. In other words, the woolly mammoth didn't evolve to have these adaptations to the cold. Rather, its predecessor already had them, meaning that these adaptations likely emerged slowly over time, rather than quickly under selective pressures. However, the woolly mammoth did continue to evolve; woolly mammoths that lived closer to the extinction had more genetic variants associated with sensing temperature than earlier woolly mammoths did, the researchers found.
Related: Wipe out: History's 7 most mysterious extinctions

How they did it
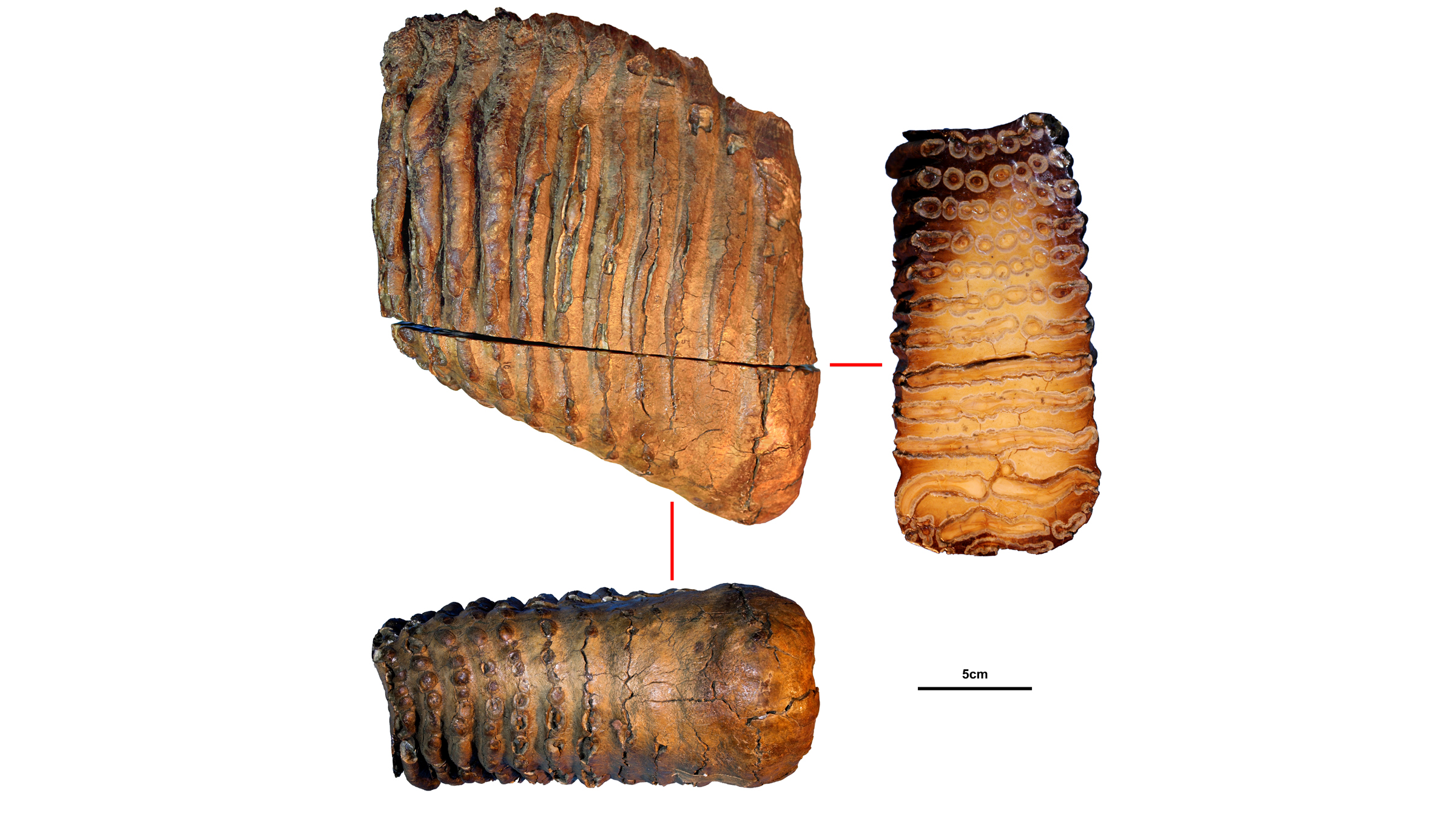
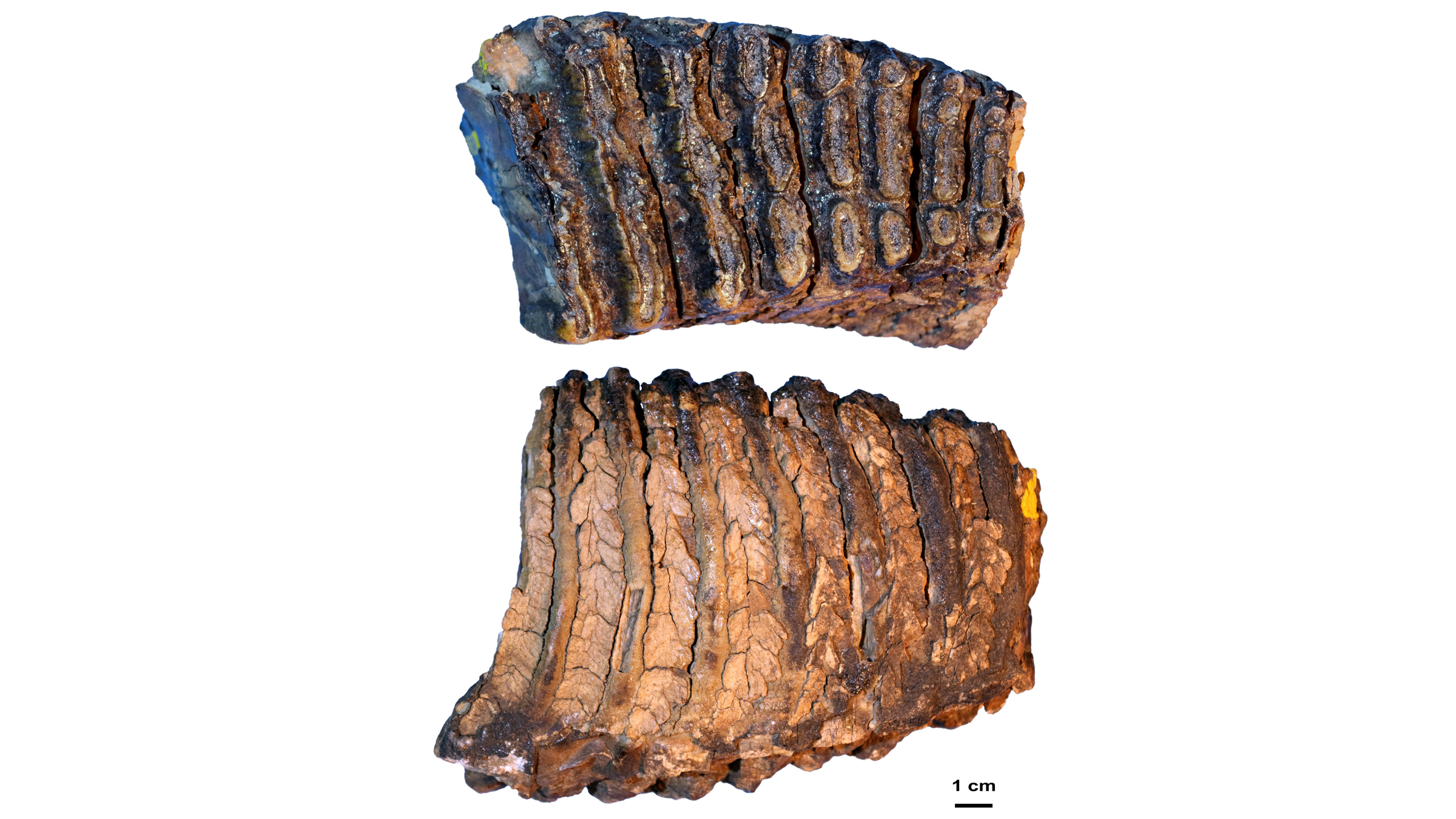
DNA starts degrading the moment an animal dies, so how did the international team of scientists piece together these three mammoth genomes? They had the already mapped the genome of the African savannah elephant (Loxodonta africana) and the mitochondrial genome (DNA inherited through the maternal line) of the Asian elephant (Elephas maximus). These previously sequenced genomes were like the image on a puzzle box; elephants are relatives of mammoths, so their genetic data served as a useful reference for the mammoths' genomes.
The tiny fragments of ancient DNA, which the team extracted from the mammoths' molar teeth, were the puzzle pieces. Though Siberia's cold permafrost helped preserve the mammoth's DNA, bacteria and other degrading elements like enzymes had broken that DNA into small fragments, some as short as 35 base pairs, the "letters" that make up DNA. Using an algorithm, the researchers discarded the foreign DNA that had contaminated the mammoth samples, including DNA from microbes. They also discarded any uncertain puzzle pieces of DNA that appeared to fit in the mammoth genome, but could have come from other animals that had contaminated the samples.
After the algorithm analyzed billions and billions of DNA puzzle pieces, the researchers had the mammoth's genomes, some more complete than others. The 1.2 million-year-old mammoth, Krestovka was the least complete at about 49 million base pairs, "which is a relatively small fraction of [the] genome, but still more than enough to confidently place the genome on the [family] tree," van der Valk said.
The 1 million-year-old steppe mammoth, Adycha, had about 884 million base pairs, or 25% to 30% of its genome completed. And the 700,000-year-old woolly mammoth, Chukochya, had about 3.6 billion base pairs, or 70% to 80% of its genome sequenced, van der Valk said. Meanwhile, the team was able to completely reassemble the mitochondrial genomes of each mammoth.
The mitochondrial DNA was key, because researchers know the rate at which mitochondrial gene mutations occur, which helped them date the samples. The team also dated the mammoths' based on geological data, such as biostratigraphy (dating the specimens based on the ages of neighboring fossils and geologic formations) and paleomagnetism (dating signatures left by Earth's changing magnetic field in the samples).
So, how long can ancient DNA survive? The scientists think that it's possible to retrieve even older genomes. "An educated guess would be that we could recover DNA that is 2 million years old, and possibly go even as far back as 2.6 million," study co-researcher Anders Götherström, a professor in molecular archaeology and joint research leader at the Center for Palaeogenetics, said in a statement. "Before that, there was no permafrost where ancient DNA could have been preserved."
The study was published online Wednesday (Feb. 16) in the journal Nature.
Originally published on Live Science.

Laura is the archaeology and Life's Little Mysteries editor at Live Science. She also reports on general science, including paleontology. Her work has appeared in The New York Times, Scholastic, Popular Science and Spectrum, a site on autism research. She has won multiple awards from the Society of Professional Journalists and the Washington Newspaper Publishers Association for her reporting at a weekly newspaper near Seattle. Laura holds a bachelor's degree in English literature and psychology from Washington University in St. Louis and a master's degree in science writing from NYU.