Ticking time bombs of DNA mutation may dictate when animals die
Animals pick up mutations at different rates.

Animals carry "mutational clocks" in their cells that dictate how quickly their DNA picks up mutations. And across species, animals tend to die once they've hit a certain number of mutations, new research finds.
It turns out that, in long-lived mammals like humans, these mutational clocks tick slower than they do in short-lived mammals like mice, meaning humans reach that threshold number of mutations at a later age than mice do. This discovery, the researchers said, could help solve a long-standing mystery in biology.
This mystery, known as Peto's paradox, describes a perplexing phenomenon that has defied explanation since the 1970s. At that time, scientists knew that animal cells accrued mutations in their DNA over time, and that as the number of mutations increased, so too did the risk of those cells turning cancerous. On paper, this suggests that the world's longest-living and largest animals should face the highest risk of cancer, because the chance of picking up cancer-causing mutations increases over time and as the total number of cells in an organism goes up.
But oddly enough, large, long-lived animals develop cancer at similar rates as tiny, short-lived creatures — this is Peto's paradox. Now, in a new study, published April 13 in the journal Nature, scientists offer a partial potential solution to this puzzle: They discovered that short- and long-lived mammals both accumulate a similar number of genetic mutations over their lifespans, but the long-lived animals do so at a far slower rate.
"I was really surprised" at the strength of the relationship between lifespan and mutation rate in different species, said Alex Cagan, a staff scientist at the Wellcome Sanger Institute in England and first author of the study. The study results help explain one aspect of Peto's paradox, by showing that having a lengthy lifespan doesn't put animals at higher risk of cancer-causing mutations. However, the authors didn't find a strong link between animals' body masses and their mutational clocks, so their results don't address the question of why big animals don't have high rates of cancer.
Related: Scientists discover 4 distinct patterns of aging
The results do support the theory that animals age, at least in part, due to the build-up of mutations in their cells over time — although the study doesn't reveal exactly how the mutations contribute to the aging process, Cagan said.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
"Based on our results, yes, you can tell a mammal is close to the end of its species' lifespan when it has [approximately] 3,200 mutations in its colonic epithelial stem cells," which was the specific population of cells that the team analyzed. "But we don't think that it's because at 3,201, the animal will drop dead from mutation overload," Cagan said. Rather, the authors think that the relationship between animals' mutational clocks and aging might be a bit more nuanced.
Ticking clocks
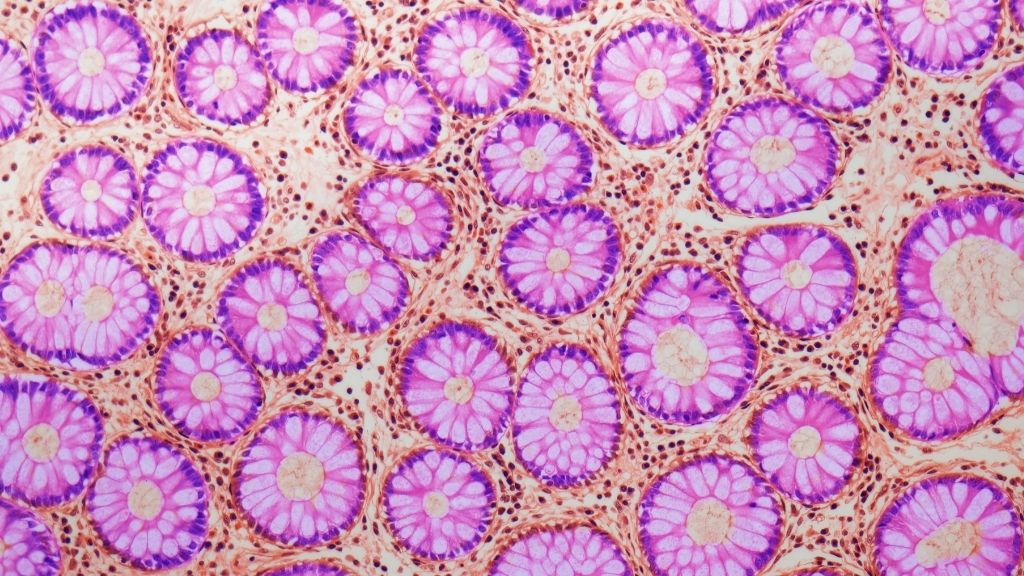
To see how quickly mutational clocks tick in different mammals, the team analyzed genetic material from 16 species: humans, black-and-white colobus monkeys, cats, cows, dogs, ferrets, giraffes, harbor porpoises, horses, lions, mice, naked mole-rats, rabbits, rats, ring-tailed lemurs and tigers. Of these species, humans have the longest lifespan at roughly 80 years; mice and rats had the shortest lifespans, between about 3 and 4 years.
From each of these species, the researchers collected DNA from "crypts," which are tiny folds found in the lining of the small intestines and colon. The cells in each crypt all descend from a single stem cell, meaning they're all clones of that stem cell. Past studies suggest that, at least in humans, crypt cells pick up mutations at a constant rate as a person ages.
In total, the researchers analyzed more than 200 crypt tissue samples from the 16 species; each sample contained a few hundred cells, Cagan noted.
"The ability to sequence the genomes of very small cell populations (e.g. those that are found within one crypt) is fairly new, so this study could not have easily been done 20 years ago," said Kamila Naxerova, an assistant professor at Harvard Medical School and a principal investigator at the Massachusetts General Hospital Center for Systems Biology, who was not involved in the study.
Related: Anti-aging vaccine shows promise in mice — will it work in humans?
The team determined the total number of DNA mutations present in each sample, and by taking each animal's age into account, they were able to estimate how quickly these mutations cropped up over the organism's lifespan. In some species, including dogs, mice and cats, the team had enough samples to compare the total number of mutations in individuals of different ages — for instance, a 1-year-old mouse versus a 2-year-old mouse — to double-check the accuracy of their mutation rate estimates.
Through their analysis, the authors discovered that, just like in humans, the crypt cells of other mammals also accrue mutations at a constant rate, year to year. But what was striking was that this mutation rate differed drastically between species. Human crypts accumulated the lowest number of new mutations each year, at only 47, while mouse crypts picked up the most, at a whopping 796 per year.
"This difference is staggering, given the large overall similarities between human and mouse genomes," Naxerov and Alexander Gorelick, a postdoctoral fellow at Harvard Medical School and Massachusetts General Hospital, wrote in an accompanying Nature commentary on the study.
Overall, the mutation rate of each species showed an inverse correlation to its lifespan, meaning that as an animal's lifespan increased the rate of new mutations per year decreased. That ultimately meant that "the total number of mutations at the end of an animal's life was roughly similar across species," Naxerova and Gorelick noted.
More mysteries to solve

The new study doesn't hint at why long-lived animals' mutational clocks tick slower than those of short-lived animals, Cagan said. That said, an earlier study, published in October 2021 in the journal Science Advances, provides one explanation.
In that study, scientists sampled fibroblasts — a type of cell found in connective tissue — from the lungs of mice, guinea pigs, blind mole-rats, naked mole-rats and humans and then exposed these cells to a mutagen, or a chemical that damages DNA. "Our reasoning was that cells from long-lived species may cope much better with a mutagen than cells from short-lived species," said Jan Vijg, a professor and chair of the Department of Genetics at the Albert Einstein College of Medicine and senior author of the Science Advances report.
And that's just what they found. "Cells from a short-lived mouse quickly accumulated a lot of mutations, while in the very long-lived naked mole-rat or human, the same dose of mutagen did not even induce any mutations," said Vijg, who was not involved in the new Nature study. This suggests that long-lived animals may be better at repairing DNA damage and preventing mutations than short-lived animals, and this may partially explain why they accumulate mutations at a slower rate.
One limitation of both recent studies is that they each included just one cell type — intestinal crypt cells or lung fibroblasts, Vijg said. That said, analyses of additional cell types would likely turn up similar results, he said. "I would expect that the findings would generalize to most other somatic cells," meaning cells that aren't eggs or sperm, Naxerova agreed.
Related: Natural rates of aging are fixed, study suggests
Cagan and his team are launching such studies into additional tissue types now. At the same time, they're moving beyond mammals to study a wide range of vertebrates and invertebrates, to see if the same relationship holds across the animal kingdom, he said. For example, the team recently got a hold of tissue samples from a super-rare Greenland shark that washed ashore in the U.K. and may have been about 100 years old at the time of its death, he said. Scientists estimate that this species can live at least up to 272 years, Live Science previously reported.
Selfish cells?
Within that research, Cagan's team hopes to reveal how the steady accumulation of mutations actually contributes to aging — assuming it does at all, Cagan said. On this front, the team has proposed a theory.
They suggest that, as all somatic cells pick up mutations over time, some of those cells will develop mutations in critical genes that would normally regulate the cells' behavior. These corrupted cells become worse at their jobs but are able to multiply more efficiently than their neighbors, the theory suggests. And as these cells take over tissues in the body, this would ultimately cause organ systems to malfunction, leading to disease and death, Cagan said.
So "it's not that every cell stops working because it's accumulated a lot of mutations," he said. Rather, problematic mutations in specific cells cause those cells to go rogue, take over tissues and crowd out all the healthier, better-functioning cells. Therefore, the mutational clock of each species likely sets the pace at which these rogue cells take over, such that "it takes a lifetime before these clonal expansions of poorly functioning cells have disrupted the tissues so much that the animal can no longer function."
Such rogue cells could be described as "selfish," since they spread to the detriment of cells around them, Naxerov and Gorelick wrote in their commentary. There's evidence from animal studies that such selfish cells can emerge in the haematopoietic system — the bodily system that makes blood — and drive disease by contributing to chronic inflammation, Naxerov told Live Science.
"It could be that selfish clones in other organs contribute to disease and aging … as well, but I think this is largely hypothetical for now," she said.
Originally published on Live Science.

Nicoletta Lanese is the health channel editor at Live Science and was previously a news editor and staff writer at the site. She holds a graduate certificate in science communication from UC Santa Cruz and degrees in neuroscience and dance from the University of Florida. Her work has appeared in The Scientist, Science News, the Mercury News, Mongabay and Stanford Medicine Magazine, among other outlets. Based in NYC, she also remains heavily involved in dance and performs in local choreographers' work.