Invisible DNA lurks everywhere in the environment — and we're on the verge of decoding its secrets
Environments are littered with the DNA of the creatures that inhabit them. Analyzing it could provide a real-time view of how our planet is changing.

There's a spa floating in the middle of Lake Erie. It has a sauna, a steam room and even a cubicle filled with snow. Upstairs, there are luxury lounges, a huge library, a curated art collection by notable artists, and a panoramic lecture theater with floor-to-ceiling windows. Passengers are busy dining, surrounded by sommeliers, in fine restaurants.
One deck below, there's a pristine, state-of-the-art laboratory full of high-tech equipment, and two multimillion-dollar submersibles can take passengers down 1,000 feet (300 meters). A team of scientists is sifting through water samples and analyzing them in real time, looking at the genetic fingerprints of plankton as it floats through the water.
The researchers on Viking's Octantis cruise ship are studying environmental DNA (eDNA) — bits of genetic material that float in the water, drift through the air, or linger in the soil. Every time a living creature passes through an environment, it sheds minuscule bits of its genetic material.
Scientists first noticed traces of this genetic material decades ago, but thanks to powerful sequencing techniques, they are now beginning to analyze eDNA to characterize food webs, reveal the locations of long-lost endangered species, and show if predators are lurking in areas where humans and wildlife are in conflict.
But the technique has one problem: It generates so much data that researchers struggle to analyze it all. Now, scientists are working to combine artificial intelligence (AI) with cutting-edge sequencing to rapidly identify changes in the types and numbers of organisms in a given ecosystem. Eventually, that information could provide a real-time view of how the planet operates — and allow us to adapt to ecological changes more quickly.
"AI's going to be able to pull out [information] in a way that our other techniques just don't have the capabilities to," said Zachary Gold, research lead of the Ocean Molecular Ecology program at the National Oceanic and Atmospheric Administration's (NOAA) Pacific Marine Environmental Laboratory. "Quicker, better, faster data allows us to do things we've never dreamt of before," he told Live Science.

A treasure trove of environmental data
The term "environmental DNA," or "eDNA," was coined in the 1980s in a study describing a technique for getting DNA from a soil sample. But it wasn't until the 2000s that fast and accurate DNA sequencing machines became widely available and affordable, making eDNA analysis practical.
Get the world’s most fascinating discoveries delivered straight to your inbox.
Next-generation sequencing (NGS) now allows scientists to analyze DNA incredibly quickly — the entire human genome can now be sequenced in just one day. For eDNA, NGS means thousands of species can be identified from a single water sample. The sequencing technology is highly advanced, but the ability to analyze and draw meaningful conclusions from it requires a huge amount of computing power and could take years of scientists' time.
The physical samples can take anywhere from a couple of days to a month to sequence, then once the sequences come back, many gigabytes of data must be downloaded and "cleaned" — that is, checked by a computer for mistakes, duplicates or formatting issues. Only then can validated datasets be analyzed.
It's that next step where AI could be transformative.
"Researchers can spend months looking through that data to try to understand and identify what are the most interesting and more powerful stories and assets that are coming out of this data, but the AI could do it, you know, in seconds," Gold said.

An army of floating laboratories
Viking began studying eDNA in part because of the pandemic. The company initially required guests to take daily polymerase chain reaction (PCR) tests for COVID-19, but once that requirement was phased out, the equipment on board its ship Octantis was repurposed to allow for real-time testing of water samples. The cruise company teamed up with NOAA in 2020, and scientists joined Viking's expedition to the Great Lakes in 2022.
Now, scientists aboard this 673-foot-long (205 m) cruise ship analyze phytoplankton in the waters they pass through, providing a snapshot of the ecosystem each time the ship visits the same regions. Compared with traditional scientific research expeditions, which are expensive and irregular, tourism vessels save time and money — cruise ships are going on these voyages anyway — and the food is a lot better, the team said.
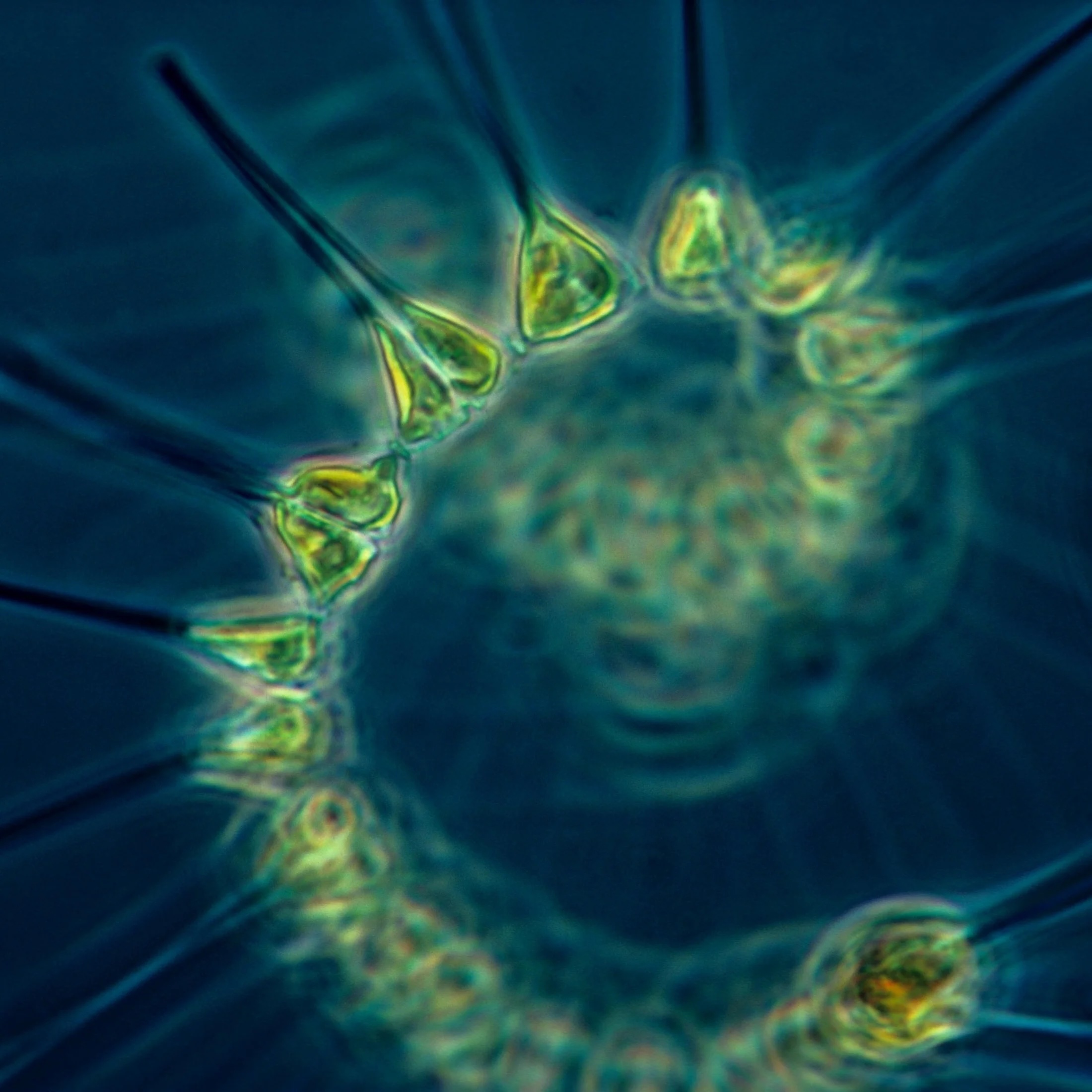
In their floating lab, researchers working with Viking now sequence phytoplankton. "They are the key to life on Earth," said Allison Cusick, a researcher at the Scripps Institution of Oceanography at the University of California, San Diego, who works in one of Viking's ship laboratories to study eDNA in remote locations like Antarctica. Phytoplankton are the foundation of most marine food webs, and they produce about half the planet's oxygen via photosynthesis. The differences among phytoplankton species is mind-blowing — the diversity between two types can be greater than that between a human and a fungus, Cusick said.
Changes in the type of plankton in the water are key indicators of biodiversity and ocean health — shifts can ricochet up the food web, with potentially devastating consequences.
Using eDNA analysis to uncover evolutionary relationships between species and the different evolutionary paths they took — for example, when one arose and when specific genes were introduced — could help scientists predict how climate change will affect different species, said Benoit Morin, a supercomputer engineer at IFREMER (the French National Institute for Ocean Science and Technology).
"By looking at the past, we can try to understand the future," Morin told Live Science.
An "Enigma project" for eDNA
To be really powerful, projects like the Viking-NOAA collaboration will need to integrate artificial intelligence into eDNA analysis.
Already, AI is being used to find potentially new species from large data sets from camera traps and automated monitoring systems. It's also being used to rediscover lost species, including the critically endangered De Winton's golden mole (Cryptochloris wintoni), which, until it was traced using eDNA, hadn't been seen for over 80 years.
But for these efforts to reach their full potential, AI techniques will need to be refined and integrated into eDNA analysis.
Once scientists have collected an eDNA sample, they analyze it via bar coding, which can either look for a single species or organism or identify multiple species at once. The barcode is a small series of unique DNA sequences that are used to identify an organism by comparing it to an online reference database.
Letizia Lamperti, a mathematical engineer at the École Pratique des Hautes Études (Practical School of Advanced Studies) in France, is developing a machine learning system to use such bar coding to reveal the health of a given environment, based on the type and number of organisms within a sample. That information, in turn, could point to potential fixes.
For example, if there was an increase in toxin-producing phytoplankton in a water sample, it may be possible to pin those changes to agricultural runoff that's feeding the phytoplankton, Cusick said.
In 2023, Lamperti and her colleagues published a study showing that neural networks — multilayered machine learning algorithms that mimic the way the human brain filters and processes information — do a better job than other statistical methods of grouping closely related organisms based on their eDNA. But just like facial recognition technology, AI will likely be better at detecting abundant species, for which there is a lot of "training" data, but less effective at spotting rarer organisms.

Several other recent studies point to the promising potential for AI in eDNA research. For instance, one study found that AI can identify 90% of unknown species in a sample, even when there aren't similar sequences from closely related organisms to use for comparison.
If AI can fulfill its potential, the shift in how we understand the environment would be monumental. Cusick likened it to Alan Turing's decryption of the Germans' Enigma code during World War II. "That's going to be transformative," she told Live Science.
"A lot of the stuff isn't hard; it's just taking the existing tools that are already out there. We've just got to point the bike in the right direction."
Zachary Gold
AI could identify newfound species on an unparalleled scale. Evolutionary relationships could be determined in the blink of an eye. Monitoring and planning for environmental changes could be transformed. For instance, by rapidly analyzing eDNA samples, AI could alert swimmers in real time to the presence of brain-eating amoebas or sharks in waterways, or forecast events like harmful algal blooms before they threaten public health — similar to how we get weather alerts on our phones now.
In theory, then, resources could be redirected quickly to resolve issues before they become a problem.
This goal is achievable, Gold said, but how long it will take will depend on the resources funneled into developing the AI to do so.
A dictionary of species
At the moment, AI is missing something important: organized volumes of good data for spotting key patterns. These data need to be put in one place as a reference database, or a dictionary of species, based on their DNA.
"We need the database of reference to perform the species identification," Lamperti told Live Science. "The problem is that we don't have it."
To identify species, AI needs to learn the key signatures, or barcodes, of individual and closely related species by training on reams and reams of data. But biodiversity datasets are not in publicly available repositories, and they're not in curated, standardized formats that can be fed into trained, bespoke AI systems. "eDNA is not AI-ready," Gold said.
In the U.S., around 40,000 eDNA samples have been collected in the past decade alone, Gold estimated, but a lot of it isn't accessible. It could be "in somebody's attic or the supplemental methods of someone's scientific paper," he said.
To draw useful conclusions to help us protect and manage the environment, AI needs to learn from a baseline database that captures biodiversity in the environments we're interested in. That's a herculean effort. "It's millions of dollars; it's tons of people's time," Gold said.
Morin is currently working on this task, but it's a slow and resource-intensive process. He and his colleagues are building a genetic "dictionary" through the ATLASea project, which aims to sequence the genomes of 4,500 marine species. This information will be deposited in an open-access database for the scientific community. IFREMER is now working with data infrastructure company NetApp to classify the mass of information being collected.
With money to develop the datasets, an AI eDNA tool could be ready "really fast," Gold said. "I have no doubt that what we're doing is not technologically difficult. It's just we're not resourcing it. If we really wanted to do this and mobilize at a scale, I have no doubt by the next Olympics in Los Angeles [in 2028], we could have the tools and resources and network set up and [be] ready to do this."
If investment and resources continue at their current pace, Gold estimated it will be a "slow trickle" and we'll get there in around 15 years. But he's optimistic the timescale could be faster. "A lot of the stuff isn't hard; it's just taking the existing tools that are already out there," Gold said. "We've just got to point the bike in the right direction."
Editor's note: This article was updated at 4.25 a.m. ET on Feb. 25 to to clarify that Viking, not external agencies, required guests to take daily PCR tests during the COVID-19 pandemic.

In Science Spotlight, Live Science takes a deeper look at emerging science and gives you the perspective you need on these advances. Our stories highlight trends in different fields, how new research is changing old ideas, and how the picture of the world we live in is being transformed thanks to science.

Hannah Osborne is the planet Earth and animals editor at Live Science. Prior to Live Science, she worked for several years at Newsweek as the science editor. Before this she was science editor at International Business Times U.K. Hannah holds a master's in journalism from Goldsmith's, University of London.
You must confirm your public display name before commenting
Please logout and then login again, you will then be prompted to enter your display name.
