New glowing molecule, invented by AI, would have taken 500 million years to evolve in nature, scientists say
An artificial intelligence model has created a new protein that researchers say would have taken 500 million years to evolve in nature — if nature were capable of producing such a thing.
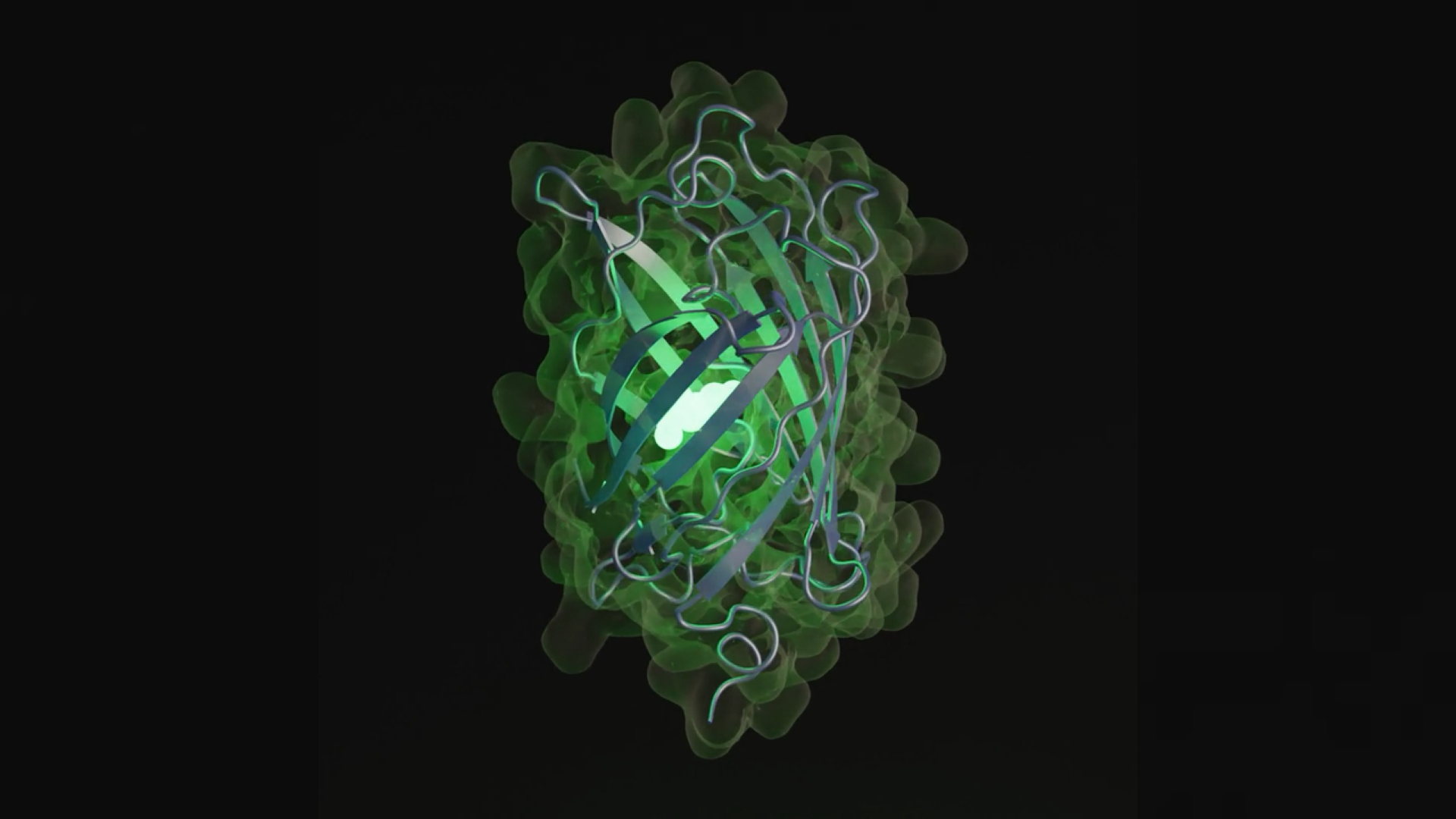
An artificial intelligence (AI) model has simulated half a billion years of molecular evolution to create the code for a previously unknown protein, according to a new study. The glowing protein, which is similar to those found in jellyfish and corals, may help in the development of new medicines, researchers say.
Proteins are one of the building blocks of life and perform various functions in the body, such as building muscles and fighting disease. The simulated protein, named esmGFP, only exists as computer code, but contains the blueprint for a previously unknown type of green fluorescent protein. In nature, green fluorescent proteins give fluorescent jellyfish and corals their glow.
The sequence of letters that spell out the instructions to make esmGFP is only 58% similar to the closest known fluorescent protein, which is a human-modified version of a protein found in bubble-tip sea anemones (Entacmaea quadricolor) — colorful sea creatures that look like they have bubbles on the ends of their tentacles. The rest of the sequence is unique, and would require a total of 96 different genetic mutations to evolve. These changes would have taken more than 500 million years to evolve naturally, according to the study.
Researchers at a company called EvolutionaryScale unveiled esmGFP and the AI model used to create it, ESM3, in a preprint study last year. Independent scientists have now peer-reviewed those findings, which were published Jan. 16 in the journal Science.
ESM3 doesn't design proteins within the usual constraints of evolution. Instead, it's a problem-solver that fills in gaps of incomplete protein code provided by the researchers, and in doing so designs something that could exist based on all of the potential pathways evolution could take.
"We've found that ESM3 learns fundamental biology, and can generate functional proteins outside the space explored by evolution," study co-author Alex Rives, co-founder and chief scientist of EvolutionaryScale, told Live Science in an email.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
The new study builds on research that Rives and his colleagues began at Meta, the parent company of Facebook and Instagram, before starting EvolutionaryScale in 2024. ESM3 is their latest version of a generative language model similar to OpenAI's GPT-4, which runs ChatGPT, but it's based on biology.
Proteins are made up of chains of molecules called amino acids, the sequence of which is provided by genes. Different proteins have different amino acid sequences. They also differ structurally, each folding into a unique shape that allows them to carry out their function, according to Nature Education. For ESM3 to understand proteins, researchers fed the model data on the main properties of a protein — amino acid sequence, structure and function — as a series of letters.
The team trained ESM3 on data from 2.78 billion proteins found in nature. The researchers then randomly hid parts of a protein blueprint and had ESM3 plug in the gaps to complete the code based on what it had learned.
"The same way a person can fill in the blanks in the soliloquy "to _ or not to _, that is the _," we can train a language model to fill in the blanks in proteins," Rives said. "Our research has shown that by solving this simple task, information about the deep structure of protein biology emerges in the network."
Scientists already modify natural proteins and engineer new ones for a variety of purposes. For example, green fluorescent proteins are used widely in research labs. Their genetic code is often added to the ends of other DNA sequences to turn the proteins that they encode green. This allows scientists to easily track proteins and cellular processes. Rives noted that ESM3's capabilities can accelerate a wide range of applications for protein engineering, including with helping to design new drugs.
Tiffany Taylor, an evolutionary biologist at the University of Bath in the U.K. who was not involved in the research, reported on the preprint version of the study for Live Science in 2024. In her analysis, Taylor wrote that AI models like ESM3 will enable innovations in protein engineering that evolution can't. However, she also noted that the researchers' claim of simulating 500 million years of evolution is focussed only on individual proteins and does not account for the many stages of natural selection that ultimately create life.
"AI-driven protein engineering is intriguing, but I can't help feeling we might be overly confident in assuming we can outsmart the intricate processes honed by millions of years of natural selection," Taylor said.

Patrick Pester is the trending news writer at Live Science. His work has appeared on other science websites, such as BBC Science Focus and Scientific American. Patrick retrained as a journalist after spending his early career working in zoos and wildlife conservation. He was awarded the Master's Excellence Scholarship to study at Cardiff University where he completed a master's degree in international journalism. He also has a second master's degree in biodiversity, evolution and conservation in action from Middlesex University London. When he isn't writing news, Patrick investigates the sale of human remains.
You must confirm your public display name before commenting
Please logout and then login again, you will then be prompted to enter your display name.